This repository contains code for the R package seismicGWAS, an implementation of the seismic computational framework for mapping genetically implicated cell types and their driver genes in complex traits and diseases, by integrating GWAS summary statistics and single cell expression data.
Citation
If you use seismicGWAS we ask that you cite our paper:
Disentangling associations between complex traits and cell types with seismic. Lai Q, Dannenfelser R, Roussarie JP, Yao V. Nature communications 16.1 (2025): 8744. DOI: 10.1038/s41467-025-63753-z
About
Integrating single-cell RNA sequencing (scRNA-seq) with Genome-Wide Association Studies (GWAS) can help reveal GWAS-associated cell types furthering our understanding of the cell-type-specific biological processes underlying complex traits and disease. In order to rapidly and accurately pinpoint associations, we develop a novel framework, seismic, which characterizes cell types using a new specificity score. As part of the seismic framework, the specific genes driving cell type-trait associations can easily be accessed and analyzed, enabling further biological insights. The following figure depicts a high level overview of this process.
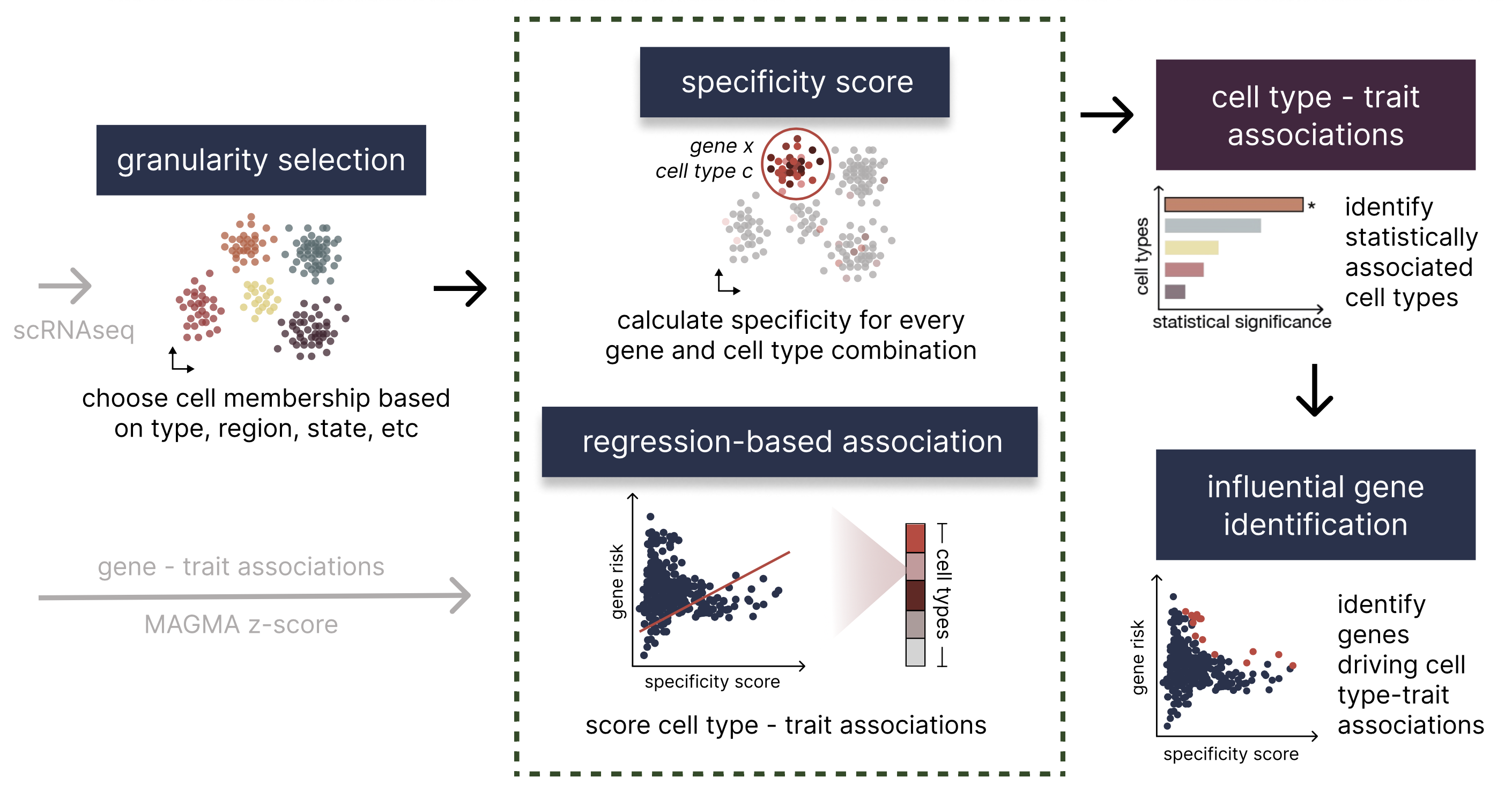
Installation and dependencies
To install the seismicGWAS R package first clone the seismic repo and then use devtools within R to point to seismic and install. The R environment version should be 4.0.0 or higher.
First download the repository to your local machine, and then directly install it in R:
devtools::install(path_to_seismic_folder)
library('seismicGWAS')Or alternatively, you can install the package directly from GitHub using the following command:
devtools::install_github("ylaboratory/seismicGWAS")
library('seismicGWAS')The package requires several dependencies which are listed in the DESCRIPTION file, which will be installed automatically.
Usage
Below we quickly show how to use seismicGWAS to calcuate cell type-trait associations for the sample data included in the package.
# calculate cell type specificity scores using included sample data
# estimated running time: less than 1 minute
tmfacs_sscore <- calc_specificity(tmfacs_sce_small, ct_label_col='cluster_name')
# convert mouse gene identifiers to human ones that match data in GWAS summary data
# from MAGMA
# estimated running time: 3 seconds
tmfacs_sscore_hsa <- translate_gene_ids(tmfacs_sscore, from='mmu_symbol')
# calculate cell type-trait associations for type 2 diabetes
# estimated running time: less than 1 minutes
get_ct_trait_associations(tmfacs_sscore_hsa, t2d_magma)
# find the influential genes for a significant cell type-trait association
# in type 2 diabetes
# estimated running time: 10 seconds
find_inf_genes("Pancreas.beta cell", tmfacs_sscore_hsa, t2d_magma)For full usage details, including a walk through of all major functions and the extra upstream data preprocessing instructions, please check out our more extensive tutorial: